Fighting COVID-19 with Deep Graph
Since December 2019, the rapid spread of COVID-19 corona-viruses worldwide has caused more than 7 million infections and more than 400,000 deaths. The rapid spread of COVID-19 demonstrates the dire need for quick and effective drug discovery. Drug repurposing is a drug discovery paradigm that uses existing drugs for new therapeutic indications. It has the advantages of significantly reducing time and cost relative to de novo drug discovery. Drug repurposing with knowledge graphs presents a promising strategy for COVID-19 treatment.
A team of AWS scientists from Amazon Shanghai AI Lab and AWS Deep Engine Science team working along with academic collaborators from the University of Minnesota, The Ohio State University, and Hunan University have created the Drug Repurposing Knowledge Graph (DRKG) and a set of machine learning tools that can be used to prioritize drugs for repurposing studies. DRKG and the ML tools are open sourced in github to help researchers conduct drug relocation research on COVID-19 and other diseases (such as Alzheimer’s disease).
DRKG is a comprehensive biological knowledge graph that relates human genes, compounds, biological processes, drug side effects, diseases and symptoms. DRKG includes, curates, and normalizes information from six publicly available databases and data that were collected from recent publications related to Covid-19. It has 97,238 entities belonging to 13 types of entities, and 5,874,261 triplets belonging to 107 types of relations.
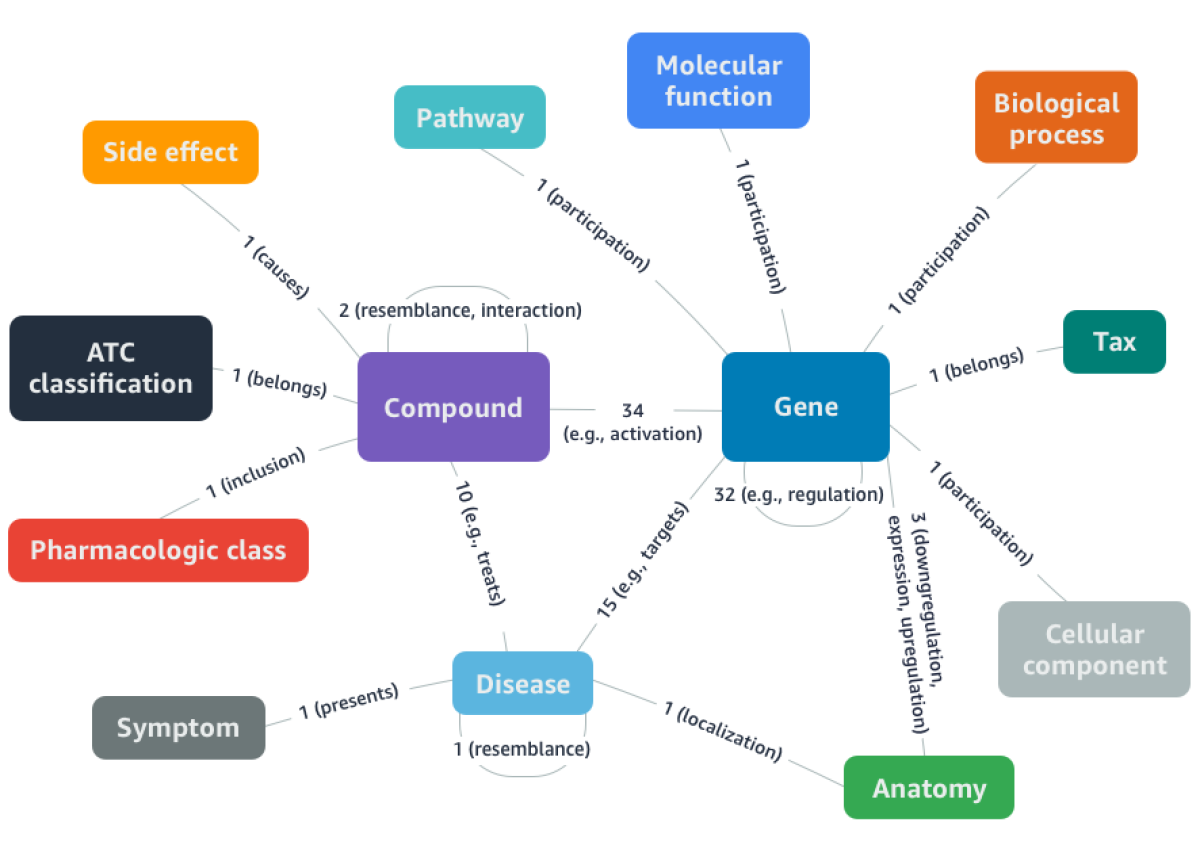
The machine learning tools use DGL-KE to learn low dimension embedding representations of entities and relations in DRKG. The resulting embeddings are used to predict how likely a drug can treat a disease or how likely a drug can bind to a protein associated with the disease. DGL-KE is a high performance, easy-to-use, and scalable package for learning large-scale knowledge graph embeddings developed by Amazon Shanghai AI Lab. The package is implemented on the top of Deep Graph Library (DGL) and developers can run DGL-KE on CPU machine, GPU machine, as well as clusters with a set of popular models, including TransE, DistMult, ComplEx, RotatE and etc. It can train a knowledge graph consisting of over 86M nodes and 338M edges in 100 minutes on an EC2 instance with 8 GPUs and 30 minutes on an EC2 cluster with 4 machines.
DRKG github repository provides examples on using DGL-KE to learn low dimension embedding representations of entities and relations in DRKG and using pre-trained knowledge graph embedding of DRKG to do drug repurposing. Preliminary experimental results show that using certain machine learning tools for COVID-19 drug discovery can identify a variety of drugs currently in clinical trials with high ranking scores.
Use DGL-KE to Learn Low Dimension Embedding Representations of Entities and Relations in DRKG
DRKG uses DGL-KE to learn knowledge graph embedding
Step 1: Downloading DRKG from here
Step 2: Load DRKG as follows.
import sys
sys.path.insert(1, '../utils')
from utils import download_and_extract
download_and_extract()
drkg_file = '../data/drkg/drkg.tsv'
Step 3: DRKG data package has a drkg.tsv file containing all triplets in the knowledge graph. Before training, We randomly divide the data set into training set, validation set and test set according to the ratio of 0.9:0.05:0.05.
import pandas as pd
import numpy as np
df = pd.read_csv(drkg_file, sep="\t")
triples = df.values.tolist()
seed = np.arange(num_triples)
np.random.shuffle(seed)
train_cnt = int(num_triples * 0.9)
valid_cnt = int(num_triples * 0.05)
train_set = seed[:train_cnt]
train_set = train_set.tolist()
valid_set = seed[train_cnt:train_cnt+valid_cnt].tolist()
test_set = seed[train_cnt+valid_cnt:].tolist()
with open("train/drkg_train.tsv", 'w+') as f:
for idx in train_set:
f.writelines("{}\t{}\t{}\n".format(triples[idx][0], triples[idx][1], triples[idx][2]))
with open("train/drkg_valid.tsv", 'w+') as f:
for idx in valid_set:
f.writelines("{}\t{}\t{}\n".format(triples[idx][0], triples[idx][1], triples[idx][2]))
with open("train/drkg_test.tsv", 'w+') as f:
for idx in test_set:
f.writelines("{}\t{}\t{}\n".format(triples[idx][0], triples[idx][1], triples[idx][2]))
Step 4: Then we directly invoke the command line toolkit provided by DGL-KE
to learn low dimension embedding representations of entities and relations in
DRKG. Here we choose the TransE_l2 model and use an AWS p3.16xlarge instance to
train the model with multi-GPU in parallel. To use other KGE model or AWS
instances please refer to DGL-KE’s documentation.
DGLBACKEND=pytorch dglke_train --dataset DRKG --data_path ./train \
--data_files drkg_train.tsv drkg_valid.tsv drkg_test.tsv \
--format 'raw_udd_hrt' --model_name TransE_l2 --batch_size 2048 \
--neg_sample_size 256 --hidden_dim 400 --gamma 12.0 --lr 0.1 \
--max_step 100000 --log_interval 1000 --batch_size_eval 16 -adv \
--regularization_coef 1.00E-07 --test --num_thread 1 \
--gpu 0 1 2 3 4 5 6 7 --num_proc 8 \
--neg_sample_size_eval 10000 --async_update
Step 5: After training, two files are generated: 1)
DRKG_TransE_l2_entity.npy, containing the low dimension embedding
representations of entities in DRKG and 2) DRKG_TransE_l2_relation.npy,
containing the low dimension embeddings representations of relations in DRKG.
These embeddings can be used in drug repurposing tasks.
node_emb = np.load('./ckpts/TransE_l2_DRKG_0/DRKG_TransE_l2_entity.npy')
relation_emb = np.load('./ckpts/TransE_l2_DRKG_0/DRKG_TransE_l2_relation.npy')
The complete example code can be found here.
Use Pre-trained Knowledge Graph Embedding for Repurposing Drugs for COVID-19 — A collaboration work from Amazon AWS AI, Hunan University, Cleveland Clinic Lerner Center for Genomic Medicine, and University of Minnesota (Repurpose Open Data to Discover Therapeutics for COVID-19 using Deep Learning) proposed a new drug repurposing methodology for COVID-19 using the combination of knowledge graph embedding and gene-set enrichment analysis method. DRKG borrows the similar idea and provides pre-trained knowledge graph embedding of DRKG for drug repurposing for COVID-19.
First of all, we define the task of finding drugs for COVID-19 using DRKG
knowledge graph as a task of predicting the possible connections between
candidate drug entities and the COVID-19 related disease entities under the
relation of 'Hetionet::CtD::Compound:Disease' and 'GNBR::T::Compound:Disease',
i.e. the treatment relationship. We select FDA approved drugs with molecular
weight larger than 250 from DRKG as drug candidates and use 34 COVID-19 virus
related entities in DRKG as target entities. Then we predict the connection
scores of all possible triplets (Drug, Treatment, Virus) under the TransE_L2
algorithm and sort the scores. Finally, we choose the top100 highest confident
connections and take the corresponding drugs as repurposed drugs. The detailed
steps are as following:
Step 1: Setting target virus entities, drug entities and treatment relations.
# COVID-19 related virus entities
COV_disease_list = ['Disease::SARS-CoV2 E','Disease::SARS-CoV2 M', ...]
# treatment relations
treatment = ['Hetionet::CtD::Compound:Disease','GNBR::T::Compound:Disease']
# candidate drugs(provided in infer_drug.tsv along with the whole DRKG dataset)
drug_list = []
with open("./infer_drug.tsv", newline='', encoding='utf-8') as csvfile:
reader = csv.DictReader(csvfile, delimiter='\t', fieldnames=['drug','ids'])
for row_val in reader:
drug_list.append(row_val['drug'])
Step 2: Get the pretrained DRKG knowledge graph embedding.
# Reading pretrained embeddings
entity_emb = np.load('../data/drkg/embed/DRKG_TransE_l2_entity.npy')
rel_emb = np.load('../data/drkg/embed/DRKG_TransE_l2_relation.npy')
drug_ids = th.tensor(drug_ids).long()
disease_ids = th.tensor(disease_ids).long()
treatment_rid = th.tensor(treatment_rid)
drug_emb = th.tensor(entity_emb[drug_ids])
treatment_embs = [th.tensor(rel_emb[rid]) for rid in treatment_rid]
Step 3: Predict the connection scores of all possible triplets (Drug, Treatment, Virus) under the TransE_L2 algorithm,the formula is as following:
, where is for head (i.e., drugs), for relation, for tail (i.e., virus) and is the constant value used in training.
import torch.nn.functional as fn
gamma=12.0
def transE_l2(head, rel, tail):
score = head + rel - tail
return gamma - th.norm(score, p=2, dim=-1)
scores_per_disease = []
dids = []
# predict the connection scores of (Drug, Treatment, Virus) triplets
# for each treatment type and combine them together.
for rid in range(len(treatment_embs)):
treatment_emb=treatment_embs[rid]
for disease_id in disease_ids:
disease_emb = entity_emb[disease_id]
score = fn.logsigmoid(transE_l2(drug_emb, treatment_emb, disease_emb))
scores_per_disease.append(score)
dids.append(drug_ids)
scores = th.cat(scores_per_disease)
Step 4: Sort the scores.
idx = th.flip(th.argsort(scores), dims=[0])
scores = scores[idx].numpy()
dids = dids[idx].numpy()
Step 5: Get the top-100 repurposed drugs.
topk=100
_, unique_indices = np.unique(dids, return_index=True)
topk_indices = np.sort(unique_indices)[:topk]
# top-100 drug Ids
proposed_dids = dids[topk_indices]
# top-100 drug scores
proposed_scores = scores[topk_indices]
Step 6: Six drugs in clinical trials appears in the top100 repurposed drugs. Their ranking and score is as following:
[0] Ribavirin -0.21416784822940826
[4] Dexamethasone -0.9984006881713867
[8] Colchicine -1.080674648284912
[16] Methylprednisolone -1.1618402004241943
[49] Oseltamivir -1.3885014057159424
[87] Deferoxamine -1.513066053390503
The complete example code can be found here.
Further read
- Check out the DRKG repository.
- Check out the announcement on Amazon Science.
- Blog in chinese.
09 June
